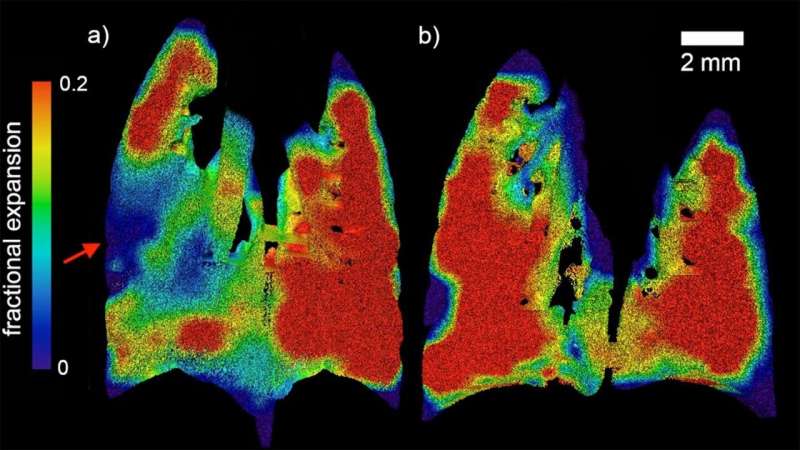
High-resolution analysis of resistant determinants and mobile elements in the human gut microbiome is quite difficult with the existing technologies. However, in the present research study, the authors highlight the effectiveness of Nanopore sequencing in getting the desired results. The analysis of genomic data derived from mobile elements in the human gut microbiome can often be limited due to the fragmented assembly from short-read sequencing.
One can overcome the limitations of fragmented assembly by using third-generation sequencing technology, which helps in getting long-reads. However, it is noted that third-generation based sequencing technologies give high-error rates and very poor throughput rates. This has resulted in limited use of technology in metagenomic related studies. The researchers in the present study have found a new way to overcome the exiting challenges by developing the first hybrid metagenomic assembler which will combine the properties of both long and short-read technologies. This will surely be considered to give high improvement when compared with the older version of assemblies along with high base par accuracy.
The approach includes a metagenome clustering technique which will be unique. It will include a scaffolding algorithm that can repeat-rich sequences with high accuracy and low error rates. Based on the numerous analysis done, the researchers identified that near-complete genomes from metagenomes can be assembled with as little as 9x long read coverage. This can enable the high-quality assembly of less abundant species. To take this understanding further, the researchers applied the concept of nanopore sequencing to analyze the gut microbiome of patients under antibiotic treatment. It was found in the study that long reads can be obtained from the samples to create accurate and efficient assemblies.